About Spl-IsoQuant
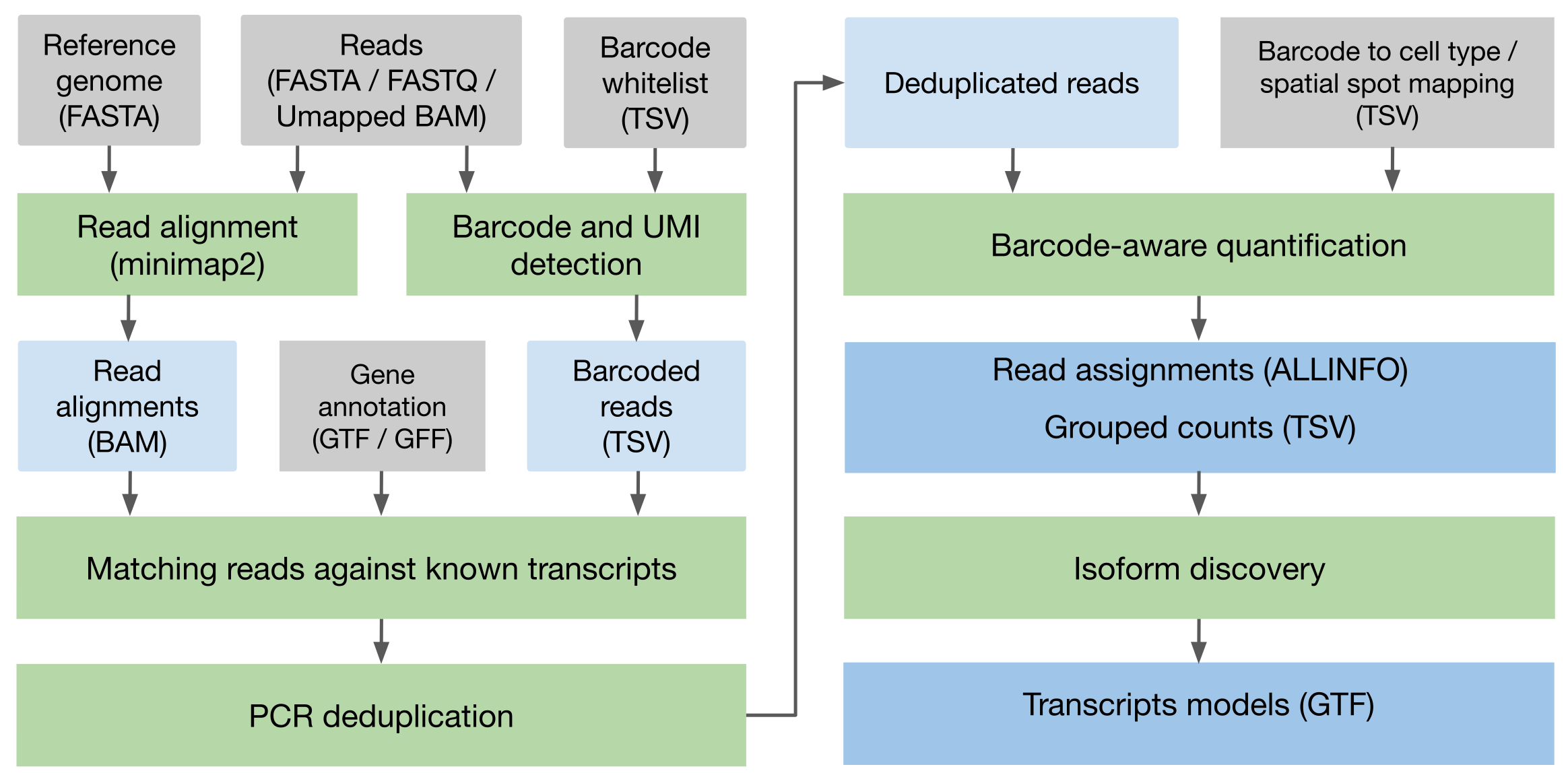
Spl-IsoQuant is a tool for single-cell and spatial long-read transcriptomics analysis. It performs genome-based analysis of long RNA reads from platforms such as PacBio or Oxford Nanopore, with specialized support for single-cell and spatial protocols. Spl-IsoQuant is capable of perfroming barcode and UMI detection for various sequencing protocols, UMI deduplication and barcode-aware quantification of reads, where reads are grouped (e.g. according to cell types or spatial location), counts are reported according to the provided grouping. We recommend providing smaller barcode whitelists (e.g. obtained from short-read sequencing) to achieve higher accuracy.
Similarly to IsoQuant, it can also perform novel transcript discovery. However, in single-cell/spatial mode, Spl-IsoQuant will only discover novel isoforms for known genes, as reads that are not assigned to any known gene are discarded during the PCR deduplication step. To achieve full transcript discovery, run Spl-IsoQuant in bulk mode.
Latest Spl-IsoQuant version can be downloaded from https://github.com/algbio/spl-IsoQuant/releases/latest.
Spl-IsoQuant pipeline